Abstract
Severe influenza A virus (IAV) infections can result in hyper-inflammation, lung injury and acute respiratory distress syndrome1,2,3,4,5 (ARDS), for which there are no effective pharmacological therapies. Necroptosis is an attractive entry point for therapeutic intervention in ARDS and related inflammatory conditions because it drives pathogenic lung inflammation and lethality during severe IAV infection6,7,8 and can potentially be targeted by receptor interacting protein kinase 3 (RIPK3) inhibitors. Here we show that a newly developed RIPK3 inhibitor, UH15-38, potently and selectively blocked IAV-triggered necroptosis in alveolar epithelial cells in vivo. UH15-38 ameliorated lung inflammation and prevented mortality following infection with laboratory-adapted and pandemic strains of IAV, without compromising antiviral adaptive immune responses or impeding viral clearance. UH15-38 displayed robust therapeutic efficacy even when administered late in the course of infection, suggesting that RIPK3 blockade may provide clinical benefit in patients with IAV-driven ARDS and other hyper-inflammatory pathologies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The single-cell RNA-sequencing datasets analysed during the current study are available in the NCBI Short Read Archive (SRA) under accession PRJNA612345. Other datasets generated during and/or analysed during the current study are PDB: 4M66, PDB: 4M69 and PDB: 7MX3. Source data are provided with this paper.
References
Kalil, A. C. & Thomas, P. G. Influenza virus-related critical illness: pathophysiology and epidemiology. Crit. Care 23, 258 (2019).
Korteweg, C. & Gu, J. Pathology, molecular biology, and pathogenesis of avian influenza A (H5N1) infection in humans. Am. J. Pathol. 172, 1155–1170 (2008).
Mauad, T. et al. Lung pathology in fatal novel human influenza A (H1N1) infection. Am. J. Respir. Crit. Care Med. 181, 72–79 (2010).
Kash, J. C. et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443, 578–581 (2006).
Flerlage, T., Boyd, D. F., Meliopoulos, V., Thomas, P. G. & Schultz-Cherry, S. Influenza virus and SARS-CoV-2: pathogenesis and host responses in the respiratory tract. Nat. Rev. Microbiol. 19, 425–441 (2021).
Nogusa, S. et al. RIPK3 activates parallel pathways of MLKL-driven necroptosis and FADD-mediated apoptosis to protect against influenza A virus. Cell Host Microbe 20, 13–24 (2016).
Rodrigue-Gervais, I. G. et al. Cellular inhibitor of apoptosis protein cIAP2 protects against pulmonary tissue necrosis during influenza virus infection to promote host survival. Cell Host Microbe 15, 23–35 (2014).
Zhang, T. et al. Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell 180, 1115–1129.e1113 (2020).
Schrauwen, E. J. & Fouchier, R. A. Host adaptation and transmission of influenza A viruses in mammals. Emerg. Microbes Infect. 3, e9 (2014).
Herfst, S., Imai, M., Kawaoka, Y. & Fouchier, R. A. Avian influenza virus transmission to mammals. Curr. Top. Microbiol. Immunol. 385, 137–155 (2014).
Sun, H. et al. Airborne transmission of human-isolated avian H3N8 influenza virus between ferrets. Cell 186, 4074–4084.e4011 (2023).
Thomas, P. G., Shubina, M. & Balachandran, S. in Curr. Top Microbiol. Immunol. Vol. 442 (eds Mocarski, E.S. & Mandal, P.) 41–63 (2020).
Balachandran, S. & Rall, G. F. Benefits and perils of necroptosis in influenza virus infection. J. Virol. 94, e01101–19 (2020).
Sanders, C. J., Doherty, P. C. & Thomas, P. G. Respiratory epithelial cells in innate immunity to influenza virus infection. Cell Tissue Res. 343, 13–21 (2011).
Sanders, C. J. et al. Compromised respiratory function in lethal influenza infection is characterized by the depletion of type I alveolar epithelial cells beyond threshold levels. Am. J. Physiol. 304, L481–L488 (2013).
Shubina, M. et al. Necroptosis restricts influenza A virus as a stand-alone cell death mechanism. J. Exp. Med. 217, e20191259 (2020).
Upton, J. W., Shubina, M. & Balachandran, S. RIPK3-driven cell death during virus infections. Immunol. Rev. 277, 90–101 (2017).
Thapa, R. J. et al. DAI senses influenza A virus genomic RNA and activates RIPK3-dependent cell death. Cell Host Microbe 20, 674–681 (2016).
Kuriakose, T. et al. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Science Immunol. 1, aag2045 (2016).
Mandal, P. et al. RIP3 induces apoptosis independent of pronecrotic kinase activity. Mol. Cell 56, 481–495 (2014).
Newton, K. et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science 343, 1357–1360 (2014).
Najjar, M. et al. Structure guided design of potent and selective ponatinib-based hybrid inhibitors for RIPK1. Cell Rep. 10, 1850–1860 (2015).
Fauster, A. et al. A cellular screen identifies ponatinib and pazopanib as inhibitors of necroptosis. Cell Death Dis. 6, e1767 (2015).
Li, J. X. et al. The B-RafV600E inhibitor dabrafenib selectively inhibits RIP3 and alleviates acetaminophen-induced liver injury. Cell Death Dis. 5, e1278 (2014).
Wissing, J. et al. Chemical proteomic analysis reveals alternative modes of action for pyrido[2,3-d]pyrimidine kinase inhibitors. Mol. Cell Proteomics 3, 1181–1193 (2004).
Meng, Y. et al. Human RIPK3 maintains MLKL in an inactive conformation prior to cell death by necroptosis. Nat. Commun. 12, 6783 (2021).
Sarhan, J. et al. Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc. Natl Acad. Sci. USA 115, E10888–E10897 (2018).
Rodriguez, D. A. et al. Caspase-8 and FADD prevent spontaneous ZBP1 expression and necroptosis. Proc. Natl Acad. Sci. USA 119, e2207240119 (2022).
Chen, J., Chen, Z., Narasaraju, T., Jin, N. & Liu, L. Isolation of highly pure alveolar epithelial type I and type II cells from rat lungs. Lab Invest. 84, 727–735 (2004).
Wang, S. & Hubmayr, R. D. Type I alveolar epithelial phenotype in primary culture. Am. J. Respir. Cell Mol. Biol. 44, 692–699 (2011).
Rishi, A. K. et al. Cloning, characterization, and development expression of a rat lung alveolar type I cell gene in embryonic endodermal and neural derivatives. Dev. Biol. 167, 294–306 (1995).
Hrdinka, M. et al. Small molecule inhibitors reveal an indispensable scaffolding role of RIPK2 in NOD2 signaling. EMBO J. 37, e99372 (2018).
Tan, Y. et al. Somatic epigenetic silencing of RIPK3 inactivates necroptosis and contributes to chemoresistance in malignant mesothelioma. Clin. Cancer Res. 27, 1200–1213 (2021).
Xie, T. et al. Structural insights into RIP3-mediated necroptotic signaling. Cell Rep. 5, 70–78 (2013).
Chen, W. et al. Diverse sequence determinants control human and mouse receptor interacting protein 3 (RIP3) and mixed lineage kinase domain-like (MLKL) interaction in necroptotic signaling. J. Biol. Chem. 288, 16247–16261 (2013).
Bantia, S. et al. Comparison of the anti-influenza virus activity of RWJ-270201 with those of oseltamivir and zanamivir. Antimicrob. Agents Chemother. 45, 1162–1167 (2001).
Groeneveld, G. H. et al. Effectiveness of oseltamivir in reduction of complications and 30-day mortality in severe seasonal influenza infection. Int. J. Antimicrob. Agents 56, 106155 (2020).
Zuliani-Alvarez, L. & Piccinini, A. M. A virological view of tenascin-C in infection. Am. J. Physiol. Cell Physiol. 324, C1–C9 (2023).
Pillay, J. et al. A subset of neutrophils in human systemic inflammation inhibits T cell responses through Mac-1. J. Clin. Invest. 122, 327–336 (2012).
Tak, T. et al. Neutrophil-mediated suppression of influenza-induced pathology requires CD11b/CD18 (MAC-1). Am. J. Respir. Cell Mol. Biol. 58, 492–499 (2018).
Nikhar, S. et al. Design of pyrido[2,3-d]pyrimidin-7-one inhibitors of receptor interacting protein kinase-2 (RIPK2) and nucleotide-binding oligomerization domain (NOD) cell signaling. Eur. J. Med. Chem. 215, 113252 (2021).
Suebsuwong, C. et al. Activation loop targeting strategy for design of receptor-interacting protein kinase 2 (RIPK2) inhibitors. Bioorg. Med. Chem. Lett. 28, 577–583 (2018).
Muraro, S. P. et al. Respiratory syncytial virus induces the classical ROS-dependent NETosis through PAD-4 and necroptosis pathways activation. Sci Rep. 8, 14166 (2018).
Wang, L. et al. Necroptosis in pulmonary diseases: a new therapeutic target. Front. Pharmacol. 12, 737129 (2021).
Ishii, K. J. et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 451, 725–729 (2008).
Murphy, J. M. et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39, 443–453 (2013).
Newton, K., Sun, X. & Dixit, V. M. Kinase RIP3 is dispensable for normal NF-κBs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Mol. Cell. Biol. 24, 1464–1469 (2004).
Berger, S. B. et al. Cutting edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J Immunol 192, 5476–5480 (2014).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Laskowski, R. A. & Swindells, M. B. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 (2011).
Boyd, D. F. et al. Exuberant fibroblast activity compromises lung function via ADAMTS4. Nature 587, 466–471 (2020).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e1821 (2019).
Rodriguez, D. A. et al. Characterization of RIPK3-mediated phosphorylation of the activation loop of MLKL during necroptosis. Cell Death Differ. 23, 76–88 (2016).
Matute-Bello, G. et al. An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am. J. Respir. Cell Mol. Biol. 44, 725–738 (2011).
Acknowledgements
We acknowledge M. Cameron, W. Childers, J. Wooltorton and K. Korzekwa for carrying out pharmacokinetic analyses of UH15-38, and L. Richey and J. Onufrak for toxicological assessment of this molecule. The authors thank J. Testa for the M29 cell line and R. Dunbrack for assistance with molecular modelling. We acknowledge The University of Texas at San Antonio Center for Innovative Drug Discovery for scale-up synthesis of UH15-38. This work was supported by NIH grants AI135025 and AI168087 to S.B.; AI144400 to S.B., G.D.C. and A.D.; AI161624 to S.B. and S.S.-C.; AI164003 to G.D.C. and A.D.; HL170121 to D.F.B. and S.B.; R01AI144828 and R35CA231620 to D.R.G.; Collaborative Influenza Vaccine Innovation Centers contract 75N93019C00052 and funds from the American Lebanese Syrian Associated Charities to S.S.-C.; HHS Contract 75N93021C00016 (St Jude Center of Excellence for Influenza Research and Surveillance) to S.S.-C. and P.G.T.; 75N93021C00018 (Center for Influenza Disease and Emergence Response) to P.G.T; and Deutsche Forschungsgemeinschaft grant SFB1160 to M. Schwemmle. Additional funds were provided by S10OD030332 to the UF Scripps Institute for Biomedical Innovation and Technology, by a Fox Chase Cancer Center Innovator Grant, and NIH Cancer Center Support Grant P30CA006927 to S.B.
Author information
Authors and Affiliations
Contributions
A.G. and D.F.B. carried out most of the biological experiments, with assistance from T.Z., I.S., L.-A.V.d.V., J.G., V.M., B.T., D.A.R., C.Y., D.S., J.B., C.D., R.M.W., M. Shubina, B.L., D.Z. and J.C.C. RIPK3 constructs, pMLKL antibodies and Casp8−/−MlklFlag/Flag mice were generated by D.A.R. and D.R.G. Test compounds were prepared by S.N., R.B. and A.L.D. Molecular docking studies were carried out by S.L. Analyses of docking results and rendering of models was performed by M.D.A. Histological analyses of infected tissues was conducted by K.Q.C. and P.V. C.L., L.C.F., M. Schwemmle, S.S.-C., D.R.G., G.D.C., P.G.T., A.D. and S.B. designed experiments and analysed the data. A.G., D.F.B., A.D., G.D.C. and S.B. wrote the manuscript. All authors participated in editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.N., G.D.C., A.D. and S.B. are listed as co-inventors on patent applications related to the UH15 series of compounds filed by Tufts University, the University of Houston and the Institute for Cancer Research, Fox Chase Cancer Center. L.C.F., G.D.C., A.D. and S.B. hold equity in Vaayu Therapeutics. G.D.C. holds equity in Denali Therapeutics and has received royalties from Brigham and Women’s Hospital. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Andrew Mehle and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 UH15-38 is a potent RIPK3 kinase inhibitor.
a, Immunoblot analysis of pMLKL and cleaved caspase 8 (CC8) in lysates obtained from Ripk3–/– MEFs stably expressing 2xFv-RIPK3 and treated with dimerizer (AP20187, 100 nM) in the presence of the pan-caspase inhibitor (IDN-6556, 20 μM) and UH15-38 (500 nM) for 12 h. b, Table comparing UH15-38 to GSK′872 in the indicated recombinant kinase assays. Kinase assays were performed using ADPGlo (Promega) and on the DiscoverX platform (Eurofins DiscoverX). ND = Not determined. c, Overview of docking of UH15-38 into human RIPK3. d, Docking of GSK′872 into mouse RIPK3. e, Docking of GSK′843 into mouse RIPK3. f, MEFs infected with PR8 (MOI = 2) were treated with indicated drugs in the presence of zVAD (50 μM) and cell viability was determined after 24 h. g, Cell survival kinetics of iBMDMs treated with LPS (10 ng/ml) and TAK1 inhibitor 5z7 (200 nM) following exposure to the indicated concentrations of the RIPK3 inhibitors UH15-38 and GSK′872 or the RIPK1 inhibitor GSK′547 for 6 h. h, Ripk3+/+ and Ripk3–/– MEFs were treated with TNF/5z7/IDN6556 and TNF/5z7, respectively, in the presence of the indicated concentrations of UH15-38, and viability was determined after 24 h. Ripk3–/– MEFs treated with UH15-38 alone were used as controls. i, Viability of Ripk3–/– MEFs treated with TNF/5z7 in the presence of DMSO or GSK′547 (10 μM). j, Immunoblot analysis of Gasdermin D (GSDMD) cleavage in primary BMDMs pre-treated with LPS (10 ng/ml) for 3 h followed by Nigericin (10 μM) or ATP (5 mM), along with DMSO or UH15-38 (500 nM), for an additional 1 h. k, Viability of primary BMDMs pre-treated with LPS (10 ng/ml for 3 h) followed by treatment with Nigericin (10 μM) and the indicated concentrations of UH15-38 for 1 h. Viability was determined by using CellTiter-Glo assay (as in g, h, k) or Trypan Blue exclusion assay (as in f). Error bars represent mean ± SD (n = 3). Data are from one of the at least two independent experiments. Groups were compared using ordinary one-way ANOVA (as in f, k) or the unpaired two-sided Student’s t test (as in i).
Extended Data Fig. 2 Pharmacological profiling of UH15-38.
a, Half-life (T1/2) and peak concentrations (Cmax) of UH15-38 in tissue samples collected from mice treated with four once-daily) doses of UH15-38 (30 mg/kg/day, i.p.). b, c, Immunofluorescence staining for phosphorylated-MLKL (pMLKL) (b) and quantification of pMLKL signal in lung, heart kidney and liver sections (c) obtained from Casp8–/–MlklFLAG/FLAG mice following i.p administration of either vehicle (n = 3 mice) or UH15-38 (30 mg/kg, once-daily, n = 4 mice) for four days. Six fields of each tissue section per mice were quantified. d, InVEST Safety Panel profile of UH15-38 showing percent inhibition of each target in the presence of 1μM compound. e, f, Immunohistochemistry for cleaved caspase 3 (CC3) (e) and quantification of cleaved caspase 3 signal (f) in lung, heart, kidney and liver sections (n = 4 mice) following i.p. administration of either vehicle or UH15-38 (30 mg/kg, once-daily) for seven days. IAV PR8-infected lung tissue (n = 4 mice) is shown as positive control in e and f (green). Groups were compared using the two-sided Mann-Whitney U test.
Extended Data Fig. 3 UH15-38 is a potent and specific inhibitor of necroptosis in murine and human cells.
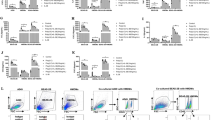
a, Podoplanin (PDPN) staining demonstrates purity of primary Type I AECs. CD140a was used as control for fibroblastic (Fibs) contamination. b, Cell survival kinetics of Type I AECs treated with TCZ and exposed to the indicated concentrations of the RIPK3 kinase inhibitors UH15-38, GSK′843 or GSK′872). c, Cell survival kinetics of primary wild-type MEFs infected with PR8 (MOI = 2) and treated with the indicated RIPK3 kinase inhibitors in the presence or absence (DMSO) of zVAD. d, Immunoblot analysis of the indicated proteins in lysates prepared from primary MEFs infected with PR8 (MOI = 2) and treated with the indicated concentrations of UH15-38. Cell lysates were prepared 12 h after infection. e, Cell survival analysis following infection of MEFs with a panel of IAV and IBV strains and exposure to the indicated concentrations of UH15-38 in the presence or absence of zVAD (50 μM). f, Immunoblot analysis of the indicated proteins in cell lysates prepared from primary MEFs infected with a panel of IAV or IBV strains (MOI = 5) and treated with DMSO or UH15-38 (1 μM). Cell lysates were prepared 12 h after infection. g, Viability of primary Ripk1–/– MEFs infected with PR8 (MOI = 2) and treated with DMSO or UH15-38 (1 μM). h, Viability of primary MEFs after infection with PR8 (MOI = 2) and treatment with UH15-38 or CSLP37 (a RIPK2 inhibitor) in the presence of either DMSO or zVAD (50 μM). (zVAD is included in the assays to block apoptosis so that effects of the test compounds on necroptosis can be evaluated). i, Viability of primary MEFs after infection with PR8 (MOI = 2) and treatment with UH15-38 or imatinib (an ABL inhibitor) in the presence of either DMSO or zVAD (50 μM). j, k, Zbp1–/– MEFs stably expressing 2xFv-tagged murine ZBP1 were exposed to dimerizer (AP20187, 100 nM) in the presence of the indicated concentrations of UH15-38. Cell viability (j) and immunoblot analyses (k) of the indicated proteins (right) are shown. l, Viability of HeLa-RIPK3 cells treated with human TNFα (100 ng/mL) + cycloheximide (2.5 μg/mL) + zVAD (50 μM) (TCZ) in the presence or absence of UH15-38 (1 μM). m, HeLa-RIPK3 cells treated with TCZ and the indicated concentrations of UH15-38 were examined for pMLKL, total MLKL, and RIPK3 by immunoblot analysis 18 h after treatment. n, M29 cells infected with PR8 (MOI = 10) were treated with increasing concentrations of UH15-38 for 24 h and examined for the indicated proteins by immunoblot analysis. H1N1 influenza strains: A/Puerto Rico/8/1934 (PR8), A/California/04/2009 (Cal/09); H3N2 influenza strains: A/Brisbane/10/2007 (Bri/07), A/Singapore/INFIMN-16-0019/2016 (Sin/16); Influenza B virus strains B/Colorado/06/2017 (Col/17) and B/Florida/04/2006 (Flo/06). zVAD (50 μM) was used to prevent apoptosis in this experiment. Cell viability in all panels was determined at 24 h after infection, using the Trypan Blue exclusion assay. Error bars represent mean ± SD of n = 3 samples. All data are from one of at least two independent experiments with similar outcomes. Groups were compared using ordinary one-way ANOVA.
Extended Data Fig. 4 UH15-38 is a potent inhibitor of RIPK3 in cellulo.
a, Viability of MEFs following treatment with indicated concentrations of UH15-38. b, Immunoblot analysis of the lysates of MEFs treated with indicated concentrations of UH15-38 for the markers of apoptosis. CC8 = cleaved caspase 8, CC3 = cleaved caspase 3. c, Viability of wild-type MEFs treated with 100 x IC50 (5 μM) of UH15-38 in the presence of DMSO or zVAD (50 μM), as compared to the viability of Ripk3 −/− MEFs treated with 100x IC50 UH15-38. d, Viability of Type I AECs following treatment with indicated concentrations of UH15-38. e, Immunoblot analysis of the lysates of Type I AECs treated with indicated concentrations of UH15-38 for markers of apoptosis. Cell viability in all panels was determined using the Trypan Blue exclusion assay. Groups were compared using ordinary one-way ANOVA.Error bars are mean ± SD of n = 3 samples. Figures are representative of two (b, e) or three (a, c, d) independent experiments with similar outcomes.
Extended Data Fig. 5 UH15-38 prevents lethality in severe influenza.
a, Weight loss curves of mice infected with PR8 (6000 EID50; ~LD100) followed by i.p. administration of vehicle (n = 20) or the following doses of UH15-38: 50 mg/kg (n = 20), 30 mg/kg (n = 21), 15 mg/kg (n = 15), 7.5 mg/kg (n = 15). Vehicle or UH15-38 was administered once-daily per the dosing schedule shown above the graph. b, c, Survival graphs (b) and weight loss curves (c) of mice (n = 10) infected with PR8 (6000 EID50) and treated with vehicle (n = 12) or with UH15-38 (30 mg/kg, i.p.) for a shortened-, or delayed-dosing regimens, as indicated above panel b. d, e, Survival (d) and weight loss (e) curves of mice (n = 7) infected with PR8 (4500 EID50) and treated i.p. with vehicle or GSK′872 (30 mg/kg) as indicated above the graph. f, Weight loss analysis of mice infected with IAV H1N1 strain A/California/04/09 (600 EID50; ~LD60) and treated once-daily with either vehicle (n = 10) or UH15-38 (30 mg/kg, i.p., n = 13) per the dosing schedule shown above the graph. g, Weight loss curves of mice infected with PR8 (4500 EID50; ~LD60) and treated i.p. with UH15-38 (30 mg/kg, i.p.), per dosing regimens shown above the graph (drug treatment beginning at day 3 after infection, n = 9; all other groups, n = 10/group). h, Wild type mice or Mlkl –/– mice (n = 10) were infected with PR8 (2500 EID50). Wild type mice were treated with either vehicle (n = 10) or 30 mg/kg UH15-38 (n = 12) once daily for four days starting 24 h after infection. i, Wild type mice or Mlkl –/– mice (n = 15) were infected with PR8 (4500 EID50). Wild type mice were treated with either vehicle (n = 14) or 30 mg/kg UH15-38 (n = 13) once daily for four days starting 24 h after infection. Mice were observed until 21 days and survival curves were plotted. Wild-type (Mlkl+/+) mice in panels h and i were generated by intercrossing Mlkl+/– mice. Groups were compared using the Log-rank (Mantel-Cox) test.
Extended Data Fig. 6 UH15-38 prevents necroptosis, inflammation, and injury in IAV-infected lungs.
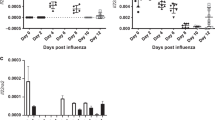
a, b, Immunofluorescence staining of cleaved caspase 3 (CC3) (a) and quantification of CC3 signal (b) in lung sections harvested on the indicated days post-infection from mice infected with PR8 (6000 EID50) and treated i.p. with either vehicle or UH15-38 (30 mg/kg once-daily), starting one day after infection, for up to four days. c, Primary Type I AECs were infected with PR8 (MOI = 2) and treated with DMSO or UH15-38 (1 μM). Supernatants were collected 12 h after infection and the indicated chemokines were analyzed on the Luminex platform. d, e, Morphometric images of Tenascin C stained lung sections showing Tenascin C positive area (red), inflamed tenascin C negative lung area (blue), larger airways (yellow) and normal lung area (green) (d) and proportion of Tenascin C positive area (e) in infected lungs (n = 5/group) nine days after infection, following i.p. administration of either vehicle or UH15-38 (30 mg/kg once-daily), starting one day after infection, for four days. f, Levels of IL-17 and CCL5 in BALF from infected mice (n = 4/group) measured on the Luminex platform three days after infection, following i.p. treatment with either vehicle or UH15-38 (30 mg/kg once-daily), starting one day after infection, for two days. g, Histological scores of lung sections obtained from mice (n = 5/group) three days (alveolar inflammation and interstitial inflammation) or nine days (septal thickening and epithelial metaplasia) following infection with PR8 (6000 EID50) and treated i.p. with either vehicle or UH15-38 (30 mg/kg once-daily), starting one day after infection, for up to four days. h, Primary BMDMs were infected with PR8 (MOI = 5) and treated with UH15-38 (1 μM) in the presence or absence of zVAD (50 μM). Supernatants were collected 24 h after infection and IL-1β and IL-18 levels were measured by ELISA. i, Airway resistance (RI) was measured in mice uninfected (Mock/Vehicle, n = 5; Mock/UH15-38, n = 4) or infected (PR8, 2500 EID50, n = 4/group) 10 days after infection following treatment with either vehicle or UH15-38 (30 mg/kg) intraperitoneally one dose daily for four days starting 24 h post infection. Data are presented as mean ± SD. (n = 6/group in b or n = 3/treatment condition in c, h). Comparison between groups was carried out by two sided Mann-Whitney U test (as in b, e, f, g) or by ordinary one-way ANOVA (as in c, h) or two-way ANOVA (as in i).
Extended Data Fig. 7 UH15-38 does not impede virus clearance or anti-IAV CD8+ T cell responses.
a,b, Lung morphometry showing virus spread (red areas) in lungs (a) and quantitation of percentage of viral antigen-positive lung area (b) at 6 days after infection. n = 5/group. c, Virus titers in lungs (day 3 and day 6, n = 10/group; day 9 and day 12, n = 5/group) at indicated time points as determined by plaque assay. d, Frequencies of IAV-specific CD8+ T cells nine days after infection with PR8 in BALF of vehicle- and UH15-38-treated mice using peptide:MHC tetramers incorporating PB1 residues 703-711 or PA residues 224-233 (n = 10/group). Comparison between groups was carried out by the two-sided Mann-Whitney U test (as in b, c, d), nd = not detected.
Extended Data Fig. 9 MLKL binding requires the displacement of the RIPK3 αC helix.
a,b, Structures of monomeric (a) and MLKL-bound (b) mRIPK3. N-terminal lobe is depicted in gold and the C-lobe in light green. The αC helix is shown in light blue, and the beginning of the activation loop, including the DFG motif, in salmon. The side chains of key active site residues are shown as sticks and labeled. c. Overlaying the monomeric and MLKL-bound conformations of mRIPK3 shows that the αC helix swivels (blue arrow) outwards in the MLKL-bound conformation, with the E61 residue now facing away from the active site. The dashed red lines indicate axes of the αC helices in inactive and active conformations.
Supplementary information
Supplementary Figure 1
Images of uncropped immunoblots
Supplementary Table 1
In vitro safety and kinome profiling of UH15-38. The first tab shows InVEST Safety Panel (Reaction Biology) results for inhibitory activity of UH15-38 (1 μM) against a panel of 50 critical enzymes. The second and third tabs depict results from screening UH15-38 (500 nM) against a panel of 90 non-mutant human kinases (KinomeScan, DiscoverX).
Supplementary Table 2
Toxicological assessment of UH15-38. The first tab depicts whole-mouse and organ weights at the end of a 7-day toxicology assessment of UH15-38, following intraperitoneal administration at 30 mg kg−1 day−1 for 7 days. The second tab shows cell composition and bloodwork at the end of the 7-day toxicology assessment. The third tab contains the Pathologist’s report.
Supplementary Table 3
Pharmacokinetic profile of UH15-38. The first tab shows key pharmacokinetic parameters of UH15-38 measured from plasma and lung levels of the compound in mice, following intraperitoneal administration of four once-daily 30 mg kg−1 doses of UH15-38. The second tab shows levels of serum biomarkers following intraperitoneal administration of four once-daily 30 mg kg−1 doses of UH15-38.
Supplementary Table 4
NMR spectra and HPLC chromatograms of UH15-38•HCl and UH15-38 Me.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gautam, A., Boyd, D.F., Nikhar, S. et al. Necroptosis blockade prevents lung injury in severe influenza. Nature 628, 835–843 (2024). https://doi.org/10.1038/s41586-024-07265-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-024-07265-8
This article is cited by
-
RIPK3 inhibitor prevents lung damage in severe influenza infection
Nature Reviews Drug Discovery (2024)
-
Blocking cell death limits lung damage and inflammation from influenza
Nature (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.